
 |
| Analysis | |
|
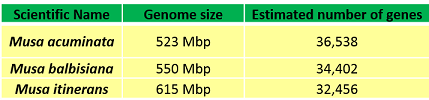
Musa has a small genome size ranging from 550 to 612 Mbp (550 Mbp M. balbisiana, 523 Mbp M. acuminata and 615 Mbp M. itenerans). The Musa acuminata reference genome sequence results from collaboration between Genoscope and CIRAD (UMR AGAP) funded by ANR. The sequence was analyzed in collaboration with several teams within the Global Musa Genomics Consortium (GMGC) and was published as version 1 (D’Hont, Angélique, et al.,2012). The reference sequence was then improved by a combination of methods and datasets, leading the release of the version 2 of the assembly and gene annotation (Martin, Guillaume, et al., 2016). | |
|
The Musa balbisiana “Pisung Klutuk Wulung” sequencing project is a collaboration between Lab. of Fruit Breeding and Biotechnology, Department of Biosystems, Katholieke Universiteit Leuven, Belgium, and The Centre for Research in Biotechnology for Agriculture and the Institute of Biological Sciences, Faculty of Science, University of Malaya, Kuala Lumpur, Malaysia (Davey, Mark W., et al., 2013). | |
|
Musa itinerans, known also as Yunnan banana, is a close relative to both banana progenitors with wide distribution across subtropical China. It is native to south-east Asia and can be found in moist ravines to mountainous areas. The genome sequencing of this species is the result of the collaboration between SCBG CAS, Bioversity International and BGI. The study was financially supported by National Natural Science Foundation of China (Wu, Wei, et al., 2016). | |
|
For SSR mining, nucleotide sequences of 11 chromosomes of M. acuminata DH Pahang v2 were downloaded from The Banana Genome Hub http://banana-genome-hub.southgreen.fr/organism/Musa/acuminata, draft sequence of M. balbisiana was downloaded from http://banana-genome-hub.southgreen.fr/organism/Musa/balbisiana, and draft sequence assembly of M. itinerans, was downloaded from http://banana-genome-hub.southgreen.fr/organism/Musa/Itiner | |
|
MIcroSAtellite identification tool (MISA) (http://pgrc.ipk-gatersleben.de/misa/) alongwith in-house developed perl scripts were used to mine the various microsatellite markers from the complete genome. Scripts were written to fragment genomic sequences, marker mining and analysing results from output. Further, experimentally validated markers were mined and curated manually from published literature and deposited in database. | |
Description of Banana genome | Frequencies of STRs based on their sizes |
 GC Content of STRs in Musa genomes 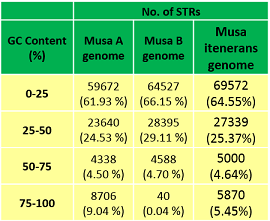 | 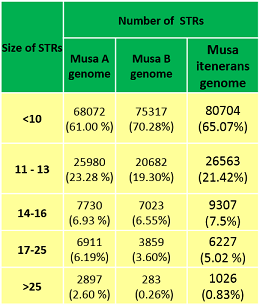 |
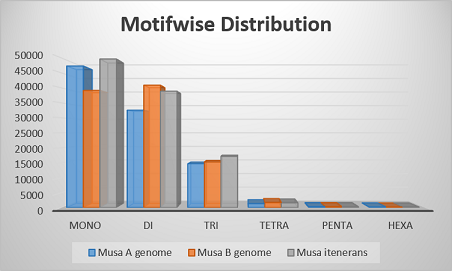
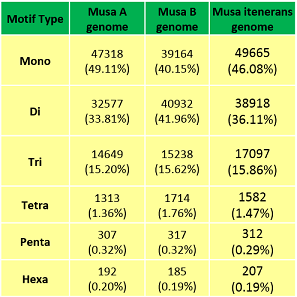
Motifwise Distribution of Banana genome | Motifwise Distribution of Banana genome |
 |  |
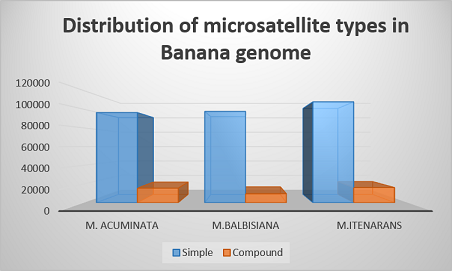
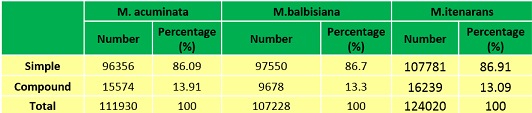
Distribution of Microsatellite Type in Banana genome | Distribution of Microsatellite Type in Banana genome |
 |  |
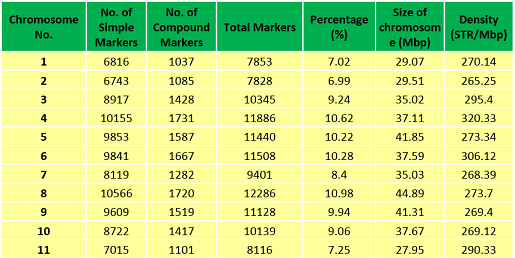
Chromosome-wide distribution of SSRs/ STRs in the Musa acuminata genome | |
 | |
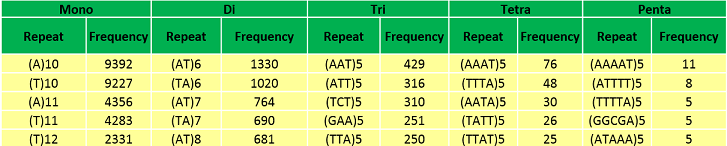
Abundant repeat types in Musa acuminata genome | |
 | |
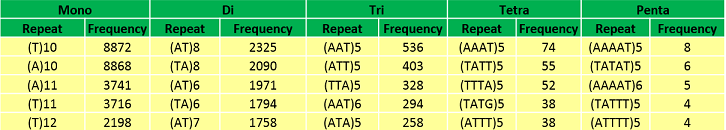
Abundant repeat types in Musa balbisiana genome | |
 | |
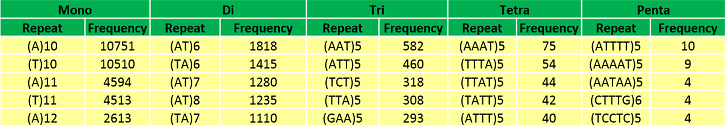
Abundant repeat types in Musa itenerans genome | |
 | |
| Copyright@2016 |